Anastomosis Groups
About the database
|
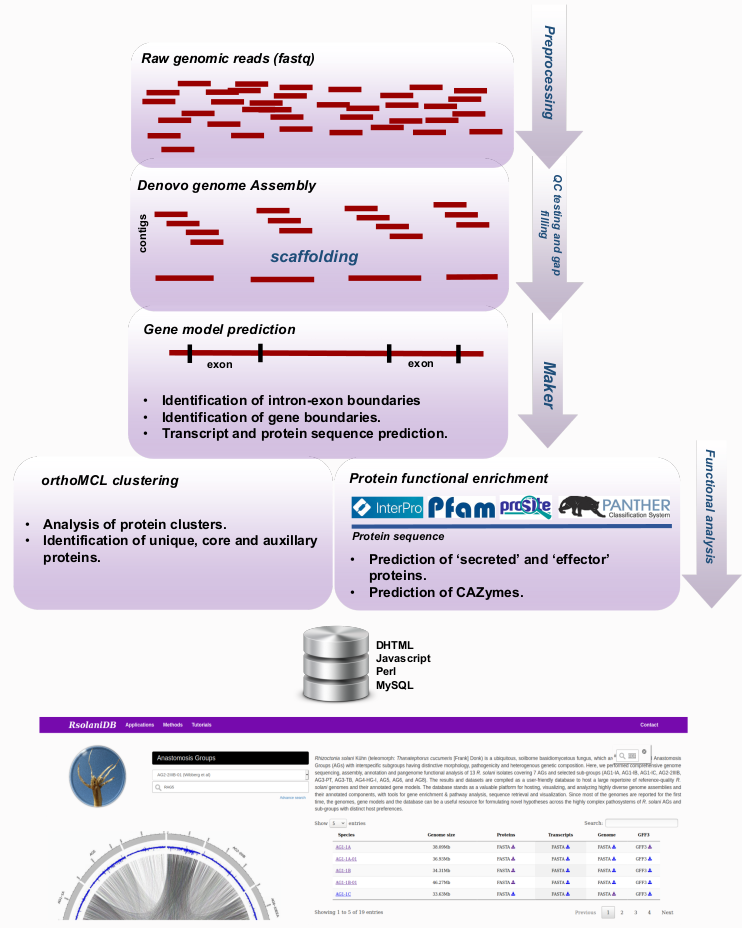
Rhizoctonia solani Kühn (teleomorph: Thanatephorus cucumeris [Frank] Donk) is a collective name for a group of ubiquitous, soilborne and multinucleate basidiomycetous fungi, which are classified into 13 Anastomosis Groups (AGs) with interspecific subgroups having distinctive morphology,
pathogenicity and heterogenous genetic composition. Here, we performed comprehensive genome sequencing, assembly, annotation and pangenome functional analysis of 12 R. solani isolates covering 7 AGs and selected sub-groups (AG1-IA, AG1-IB, AG1-IC, AG2-2IIIB, AG3-PT, AG3-TB, AG4-HG-I, AG5, AG6, and AG8).
The results and datasets are compiled as a user-friendly database to host a large repertoire of reference-quality R. solani genomes and their annotated gene models. The genomes of six previously reported R. solani isolates by other research groups are also included in this database. The database stands
as a valuable platform for hosting, visualizing, and analyzing highly diverse genome assemblies and their annotated components, with tools for gene enrichment & pathway analysis, sequence retrieval and visualization. Since most of the genomes are reported for the first time, the genomes, gene models and
the database can be a useful resource for formulating novel hypotheses across the highly complex pathosystems of R. solani AGs and sub-groups with distinct host preferences. |
Rhizoctonia diseases ⓘClick thumbnail to enlarge image
Analysis tools
Upload RsolaniDB ID(s) to retrieve its informationⓘUpload a list of RsolaniDB IDs for which information has to be downloaded. The input is the list of IDs in RsolaniDB format. The test (sample) files can be loaded with "load sample".
RDB ID? The identifier format (i.e. RDB ID) for each entry in RsolaniDB database start with ‘RS_’ and AG subgroup name followed by a unique number. We also included five previously published R. solani annotated genome sequences (i.e. AG1-1A, AG1-1B, AG2-2IIIB, AG3-PT and AG8) with their identifiers converted into the RDB ID format.
Load Sample #1
If you use RsolaniDB, please cite:
Kaushik, A., Roberts, D.P., Ramaprasad A., Mfarrej, S., Mridul Nair, Lakshman, D.K.* and Pain, A.* Pangenome Analysis of the Soilborne Fungal Phytopathogen Rhizoctonia solani and Development of a Comprehensive Web Resource RsolaniDB: Front. Microbiol., 25 March 2022; https://doi.org/10.3389/fmicb.2022.839524